Chapter 5 Pathway分析
5.1 Pathway Enrichment Analysis
KEGG通路分析,包括基因和代谢物的扩展通路分析.
dat_increase <- result_mlimma_all %>%
dplyr::filter(logFC > 0.58) %>%
dplyr::filter(P.Value < 0.05)
dat_decrease <- result_mlimma_all %>%
dplyr::filter(logFC < -0.58) %>%
dplyr::filter(P.Value < 0.05)
kegg_all <- unique(c(dat_increase$name,dat_decrease$name))
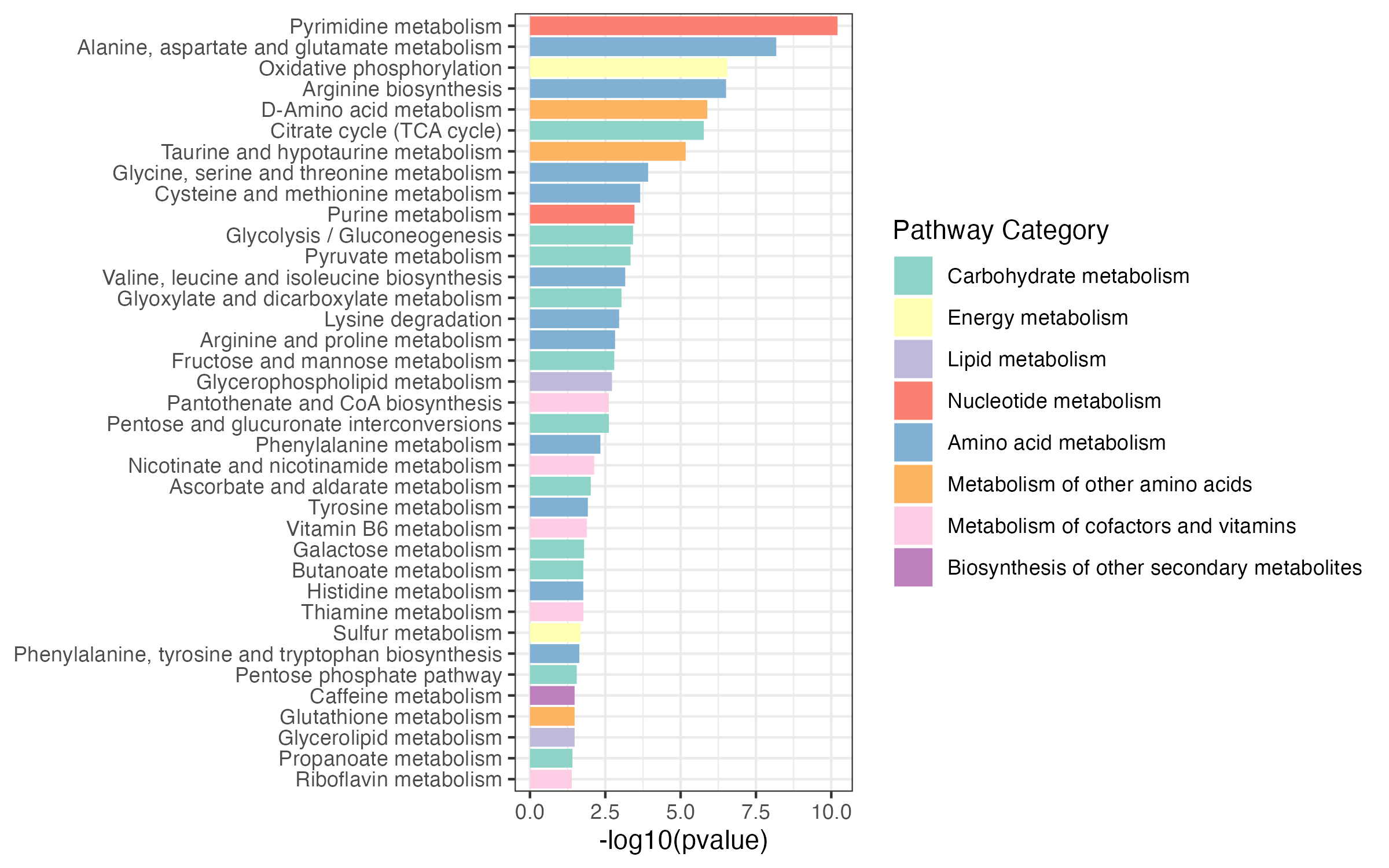
pathway_result <- PathwayAnalysis(kegg_all,out="metabolite")
ggsave("result/04.pathway_enrichment.pdf",pathway_result$p_barplot,width = 8,height = 5)
ggsave("result/04.pathway_enrichment.png",pathway_result$p_barplot,width = 8,height = 5)pathway_enrichment
5.2 Differential Abundance Analysis

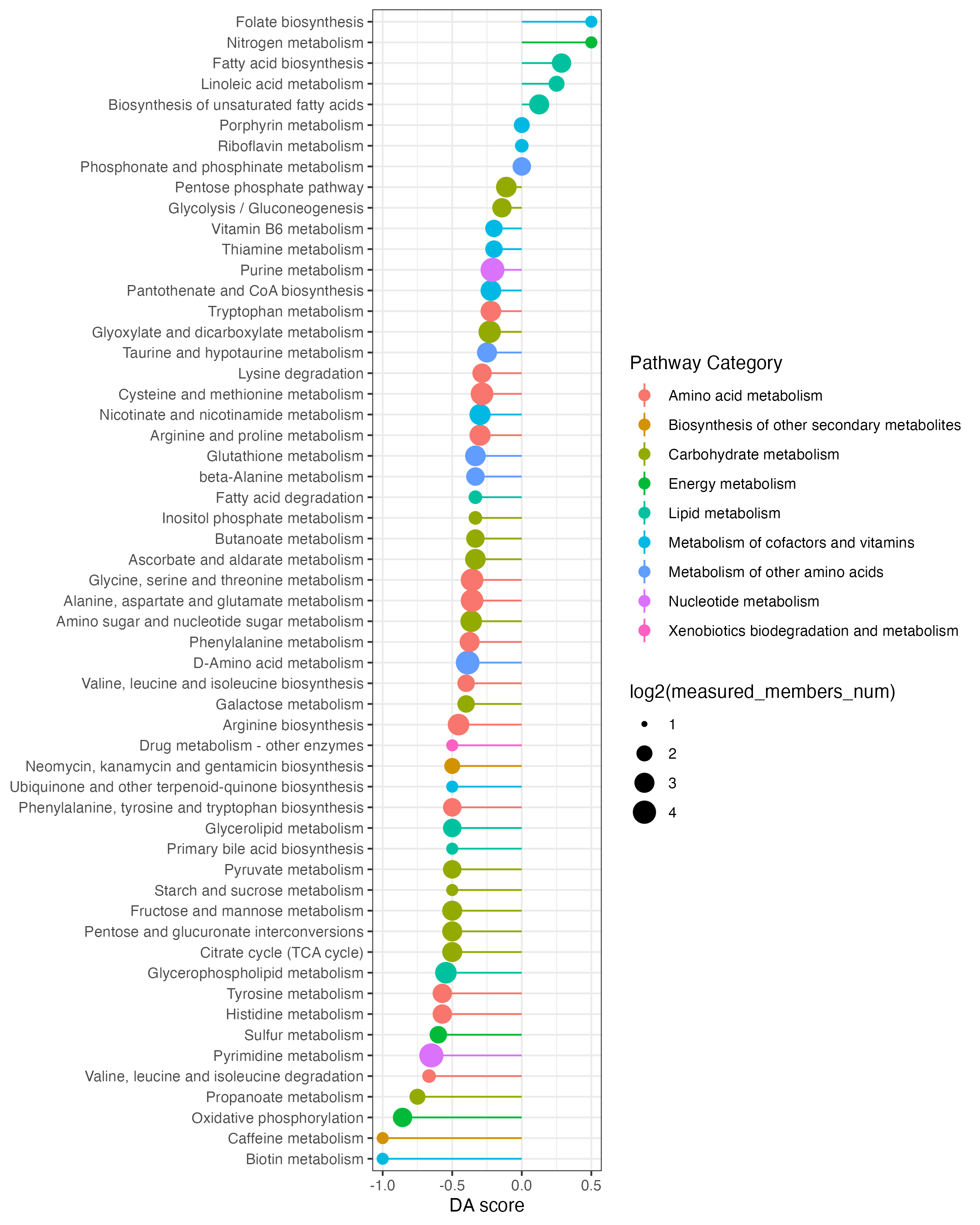
result_da <- DAscore(dat_increase$name,dat_decrease$name,result_mlimma_all$name,out="metabolite" )
View(result_da$result)
ggsave("result/04.DA_DA-score.pdf",result_da$p,width = 8,height = 10)
ggsave("result/04.DA_DA-score.png",result_da$p,width = 8,height = 10)
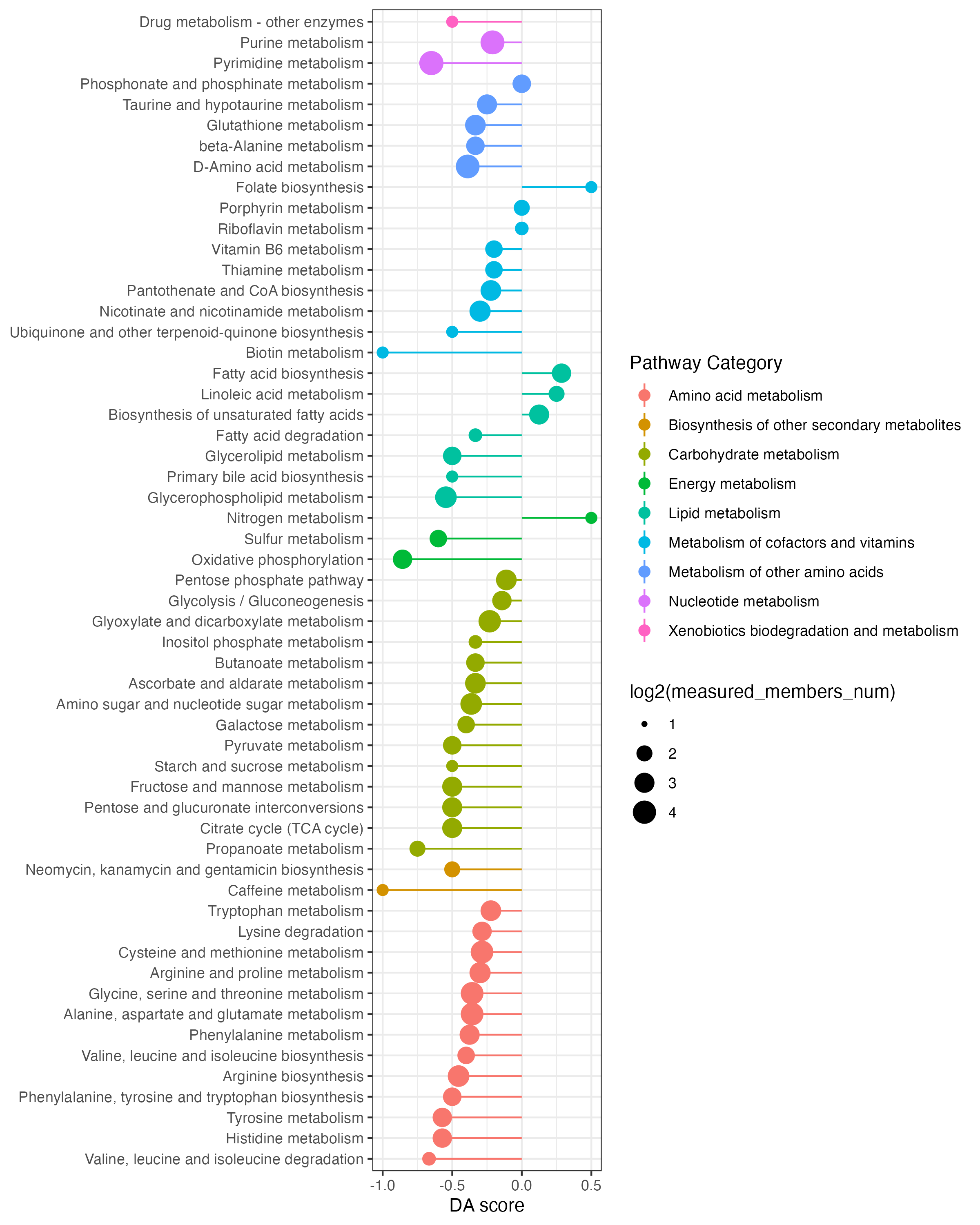
result_da2 <- DAscore(dat_increase$name,dat_decrease$name,result_mlimma_all$name,sort_plot = "category",out="metabolite" )
ggsave("result/04.DA_pathway-category.pdf",result_da2$p,width = 8,height = 10)
ggsave("result/04.DA_pathway-category.png",result_da2$p,width = 8,height = 10)DA_DA-score
DA_pathway-category
5.3 MSEA
Metabolite Set Enrichment Analysis
dd_arrange <- result_mlimma_all %>%
dplyr::arrange(logFC)
d_logFC <- dd_arrange$logFC
names(d_logFC) <- dd_arrange$name
result_msea <- MSEA(d_logFC)
result_msea <- result_msea %>%
dplyr::mutate(leadingEdge=as.character(leadingEdge))
write.table(result_msea,"result/04.MSEA.txt",quote=F,row.names=F,sep="\t")
## 想看哪个通路就输入哪个通路名字
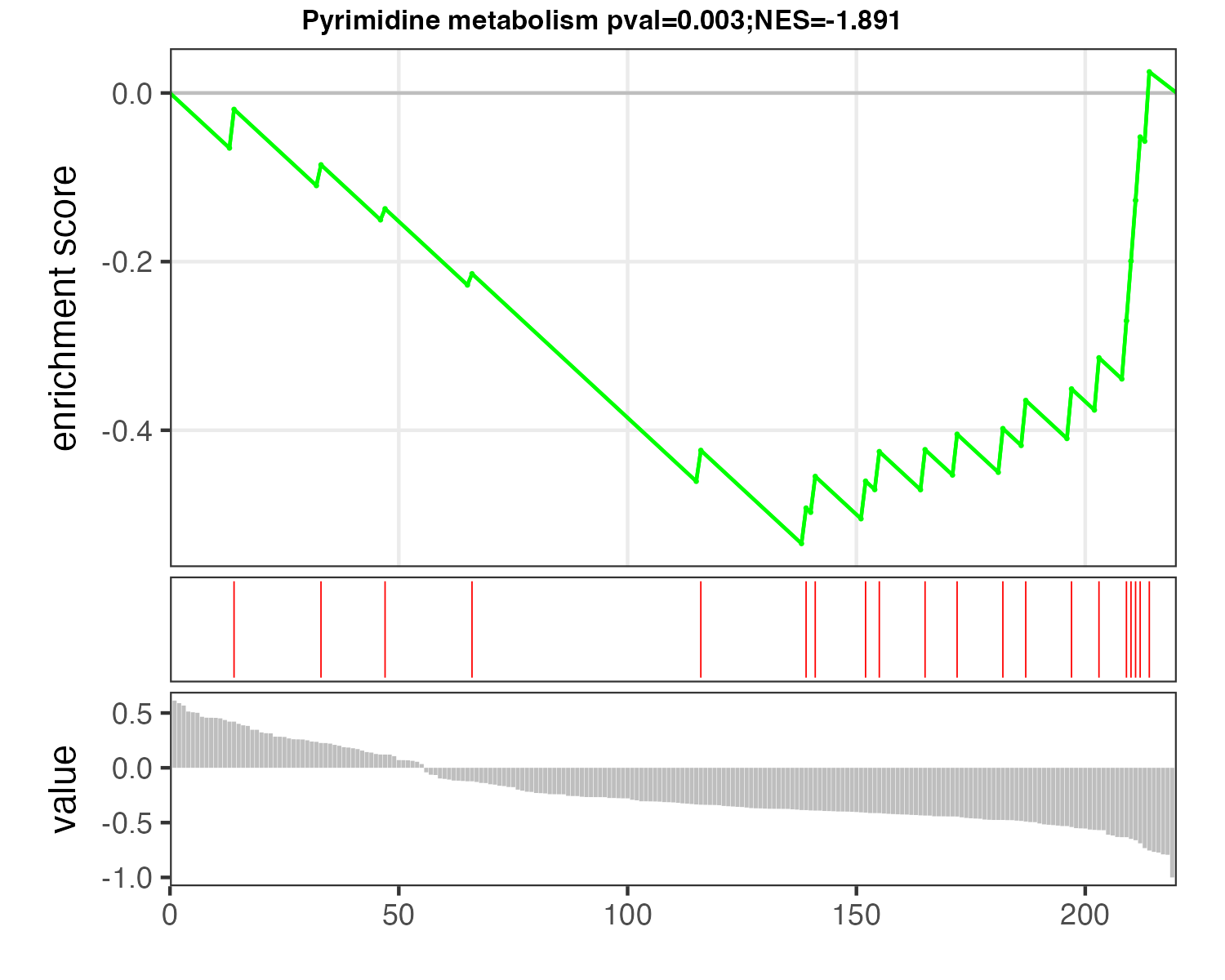
p_MSEA <- pMSEA("Pyrimidine metabolism",d_logFC)
ggsave("result/04.p_MSEA_pyrimidine.pdf",p_MSEA,width=5,height = 4)
ggsave("result/04.p_MSEA_pyrimidine.png",p_MSEA,width=5,height = 4)
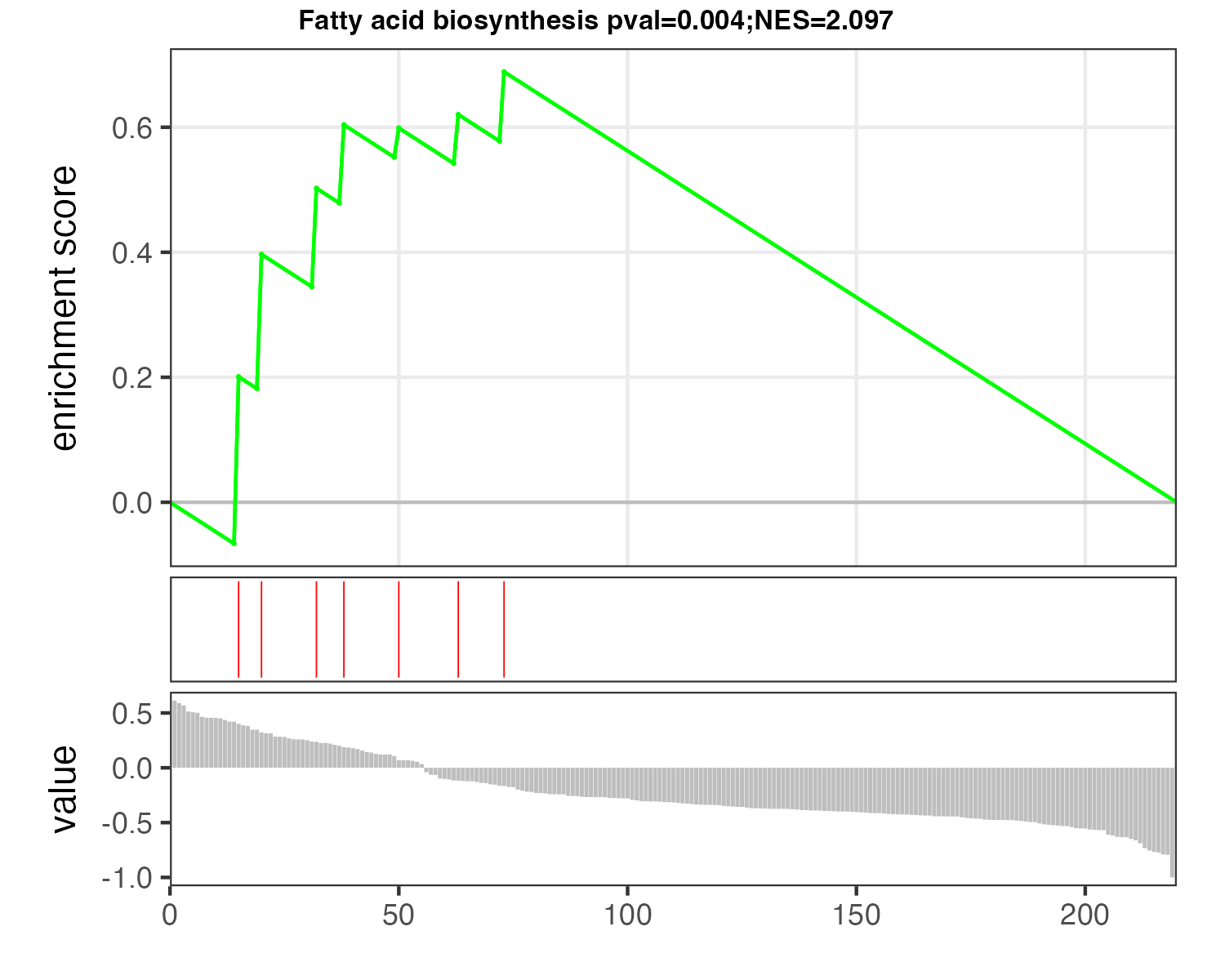
p_MSEA <- pMSEA("Fatty acid biosynthesis",d_logFC)
ggsave("result/04.p_MSEA_fatty.pdf",p_MSEA,width=5,height = 4)
ggsave("result/04.p_MSEA_fatty.png",p_MSEA,width=5,height = 4)p_MSEA_pyrimidine
p_MSEA_fatty