Chapter 4 scRNA-seq
4.1 Load scRNA-seq data
In the tutorial, the scRNA-seq of one APL (AML with PML::RARA fusion, also called acute promyelocytic leukemia)) patient (including 3,000 cells) was used for illustration.
The expample data of APL scRNA-seq can be downloaded via APL scrna demo data
# Library packages
library(Seurat)
library(HematoMap)
# Read count matrix of APL
aml.count <- readRDS("data/AML_scRNA_count.rds")
dim(aml.count)## [1] 36601 3000# Construct Seurat object
aml <- Seurat::CreateSeuratObject(counts = aml.count,
project = "aml",
min.cells = 0,
min.features = 0)## Warning: Data is of class matrix. Coercing to dgCMatrix.## An object of class Seurat
## 36601 features across 3000 samples within 1 assay
## Active assay: RNA (36601 features, 0 variable features)
## 1 layer present: counts4.2 Calculate similarities
aml <- readRDS("data/AML_scRNA_seurat.rds")
# Construct HematoMap object
hmap <- HematoMap::CreateSubclusterObject(aml)## [INFO] 2024-07-22 10:38:20.557213: Subcluster Computing##
| | 0%, ETA NA
|========= | 7%, ETA 00:58
|================== | 14%, ETA 00:48
|=========================== | 21%, ETA 00:41
|==================================== | 29%, ETA 00:36
|============================================= | 36%, ETA 00:31
|====================================================== | 43%, ETA 00:26
|================================================================ | 50%, ETA 00:22
|========================================================================= | 57%, ETA 00:19
|================================================================================== | 64%, ETA 00:15
|============================================================================================================= | 86%, ETA 00:04## Warning in irlba(A = t(x = object), nv = npcs, ...): You're computing too large a percentage of total singular values, use a standard svd instead.## [INFO] 2024-07-22 10:38:46.999342: Subcluster Complete!##
| | 0%, ETA NA
|== | 2%, ETA 00:12
|===== | 4%, ETA 00:08
|======= | 6%, ETA 00:07
|========= | 7%, ETA 00:07
|============ | 9%, ETA 00:06
|============== | 11%, ETA 00:06
|================ | 13%, ETA 00:06
|=================== | 15%, ETA 00:05
|===================== | 17%, ETA 00:05
|======================== | 19%, ETA 00:05
|========================== | 20%, ETA 00:05
|============================ | 22%, ETA 00:05
|=============================== | 24%, ETA 00:05
|================================= | 26%, ETA 00:04
|=================================== | 28%, ETA 00:04
|====================================== | 30%, ETA 00:04
|======================================== | 31%, ETA 00:04
|========================================== | 33%, ETA 00:04
|============================================= | 35%, ETA 00:04
|=============================================== | 37%, ETA 00:04
|================================================= | 39%, ETA 00:04
|==================================================== | 41%, ETA 00:04
|====================================================== | 43%, ETA 00:04
|======================================================== | 44%, ETA 00:04
|=========================================================== | 46%, ETA 00:04
|============================================================= | 48%, ETA 00:03
|================================================================ | 50%, ETA 00:03
|================================================================== | 52%, ETA 00:03
|==================================================================== | 54%, ETA 00:03
|======================================================================= | 56%, ETA 00:03
|========================================================================= | 57%, ETA 00:03
|=========================================================================== | 59%, ETA 00:03
|============================================================================== | 61%, ETA 00:02
|================================================================================ | 63%, ETA 00:02
|================================================================================== | 65%, ETA 00:02
|===================================================================================== | 67%, ETA 00:02
|======================================================================================= | 69%, ETA 00:02
|========================================================================================= | 70%, ETA 00:02
|============================================================================================ | 72%, ETA 00:02
|============================================================================================== | 74%, ETA 00:02
|================================================================================================ | 76%, ETA 00:02
|=================================================================================================== | 78%, ETA 00:01
|===================================================================================================== | 80%, ETA 00:01
|======================================================================================================= | 81%, ETA 00:01
|========================================================================================================== | 83%, ETA 00:01
|============================================================================================================ | 85%, ETA 00:01
|=============================================================================================================== | 87%, ETA 00:01
|================================================================================================================= | 89%, ETA 00:01
|=================================================================================================================== | 91%, ETA 00:01
|====================================================================================================================== | 93%, ETA 00:00
|======================================================================================================================== | 94%, ETA 00:00
|========================================================================================================================== | 96%, ETA 00:00
|============================================================================================================================= | 98%, ETA 00:00
|===========================================================================================================================| 100%, Elapsed 00:06## [INFO] 2024-07-22 10:38:54.000281: CosineDist Running## [INFO] 2024-07-22 10:39:18.238693: CosineDist Complete!## An object of class HematoSubCluster
## 54 sub-clusters with cosine distance# C0:0001 was one of the sub-clusters in hmap
# This plot illustrate the "Like" score between sub-cluster in APL and normal BMMCs
plotThetaLike(hemato.subc = hmap, plot.scell = "C0:0001")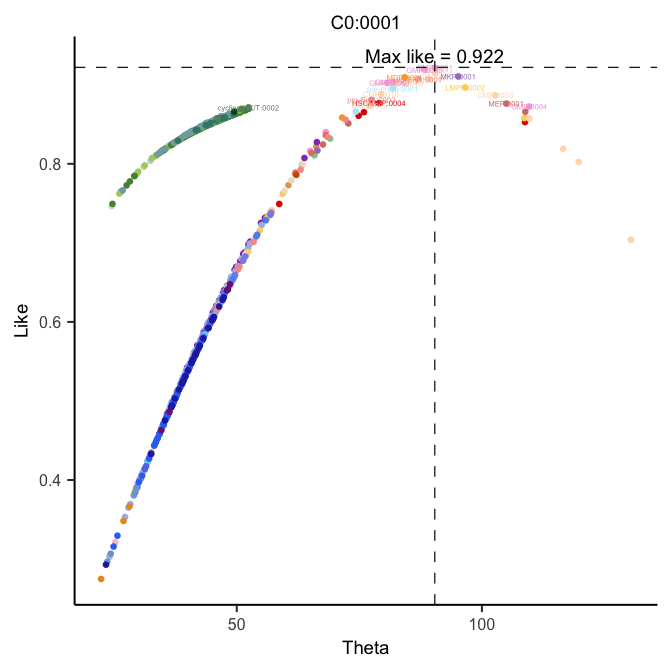
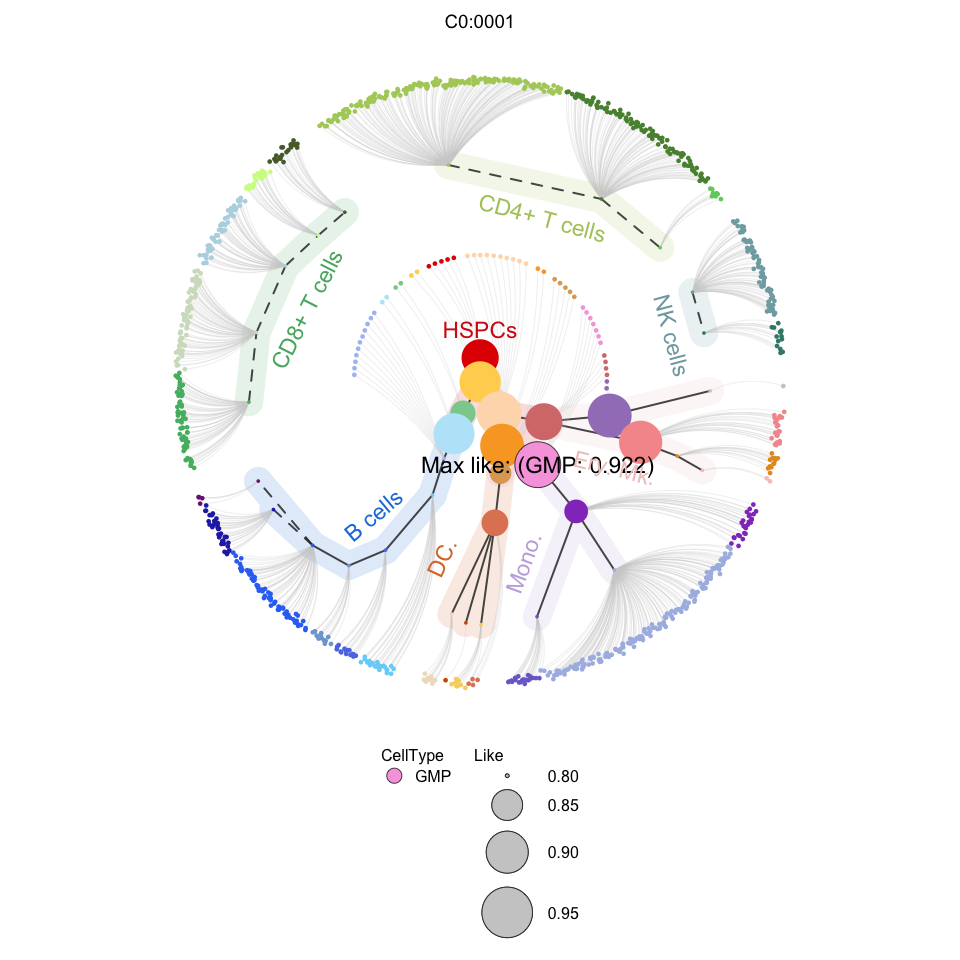
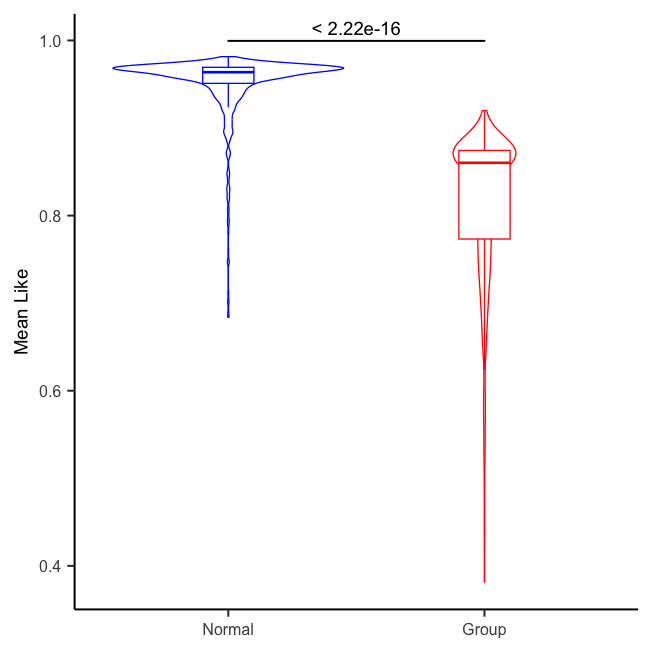
4.3 Visualization
# Visualization for APL (circle tree)
plotCircleTree(hemato.subc = hmap, group.subc = "external", top.subc = 3,
color.mapping = "cell.type", title = "APL BMMCs")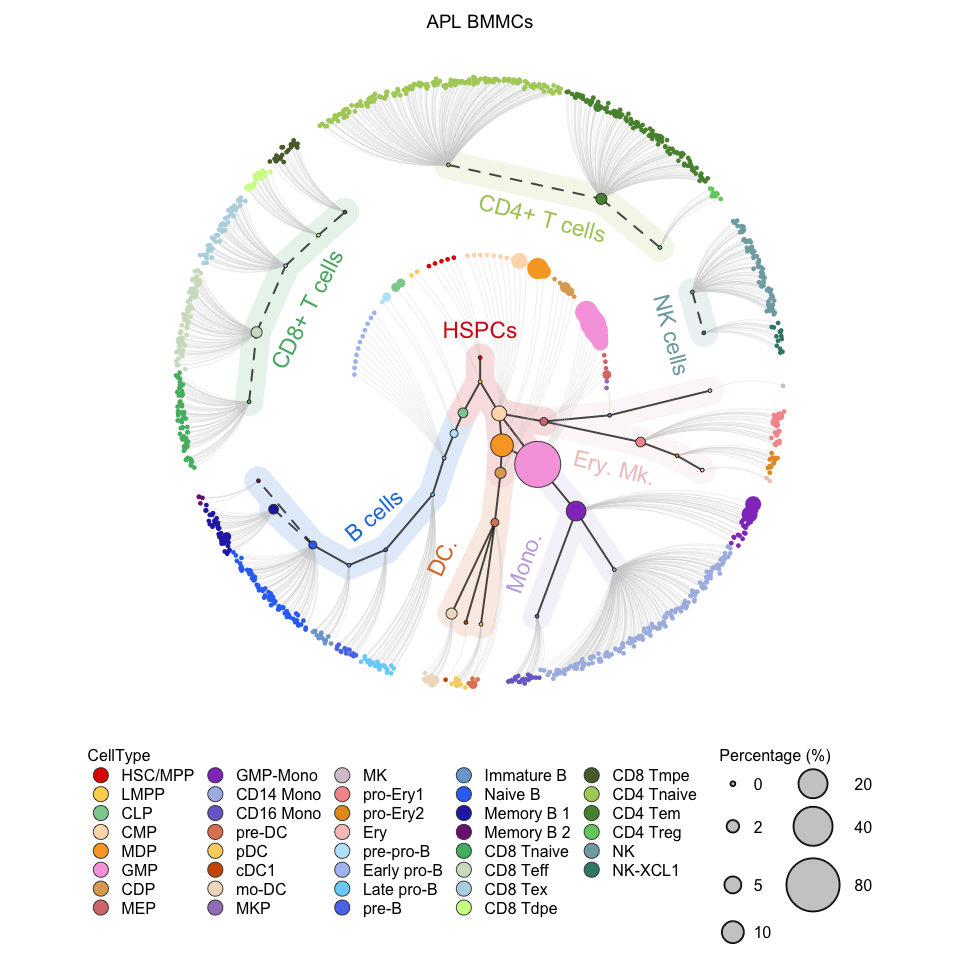
# Visualization for APL (circle tree without population label)
# clean version
plotCircleTree(hemato.subc = hmap, group.subc = "external", top.subc = 3,
color.mapping = "cell.type", title = "APL BMMCs",
label.population = FALSE)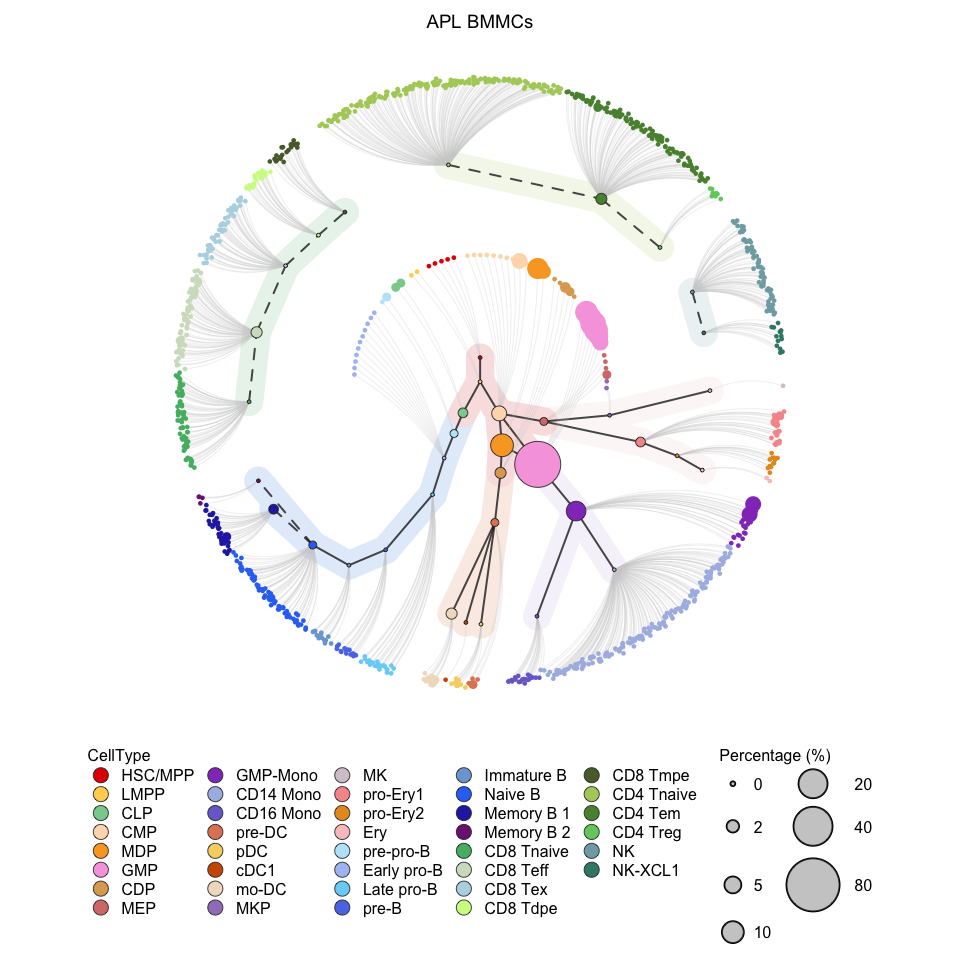
# Visualization for the normal BMMC (the reference) (circle tree)
plotCircleTree(hemato.subc = hmap, group.subc = "reference", top.subc = 3,
color.mapping = "cell.type", title = "Normal BMMCs")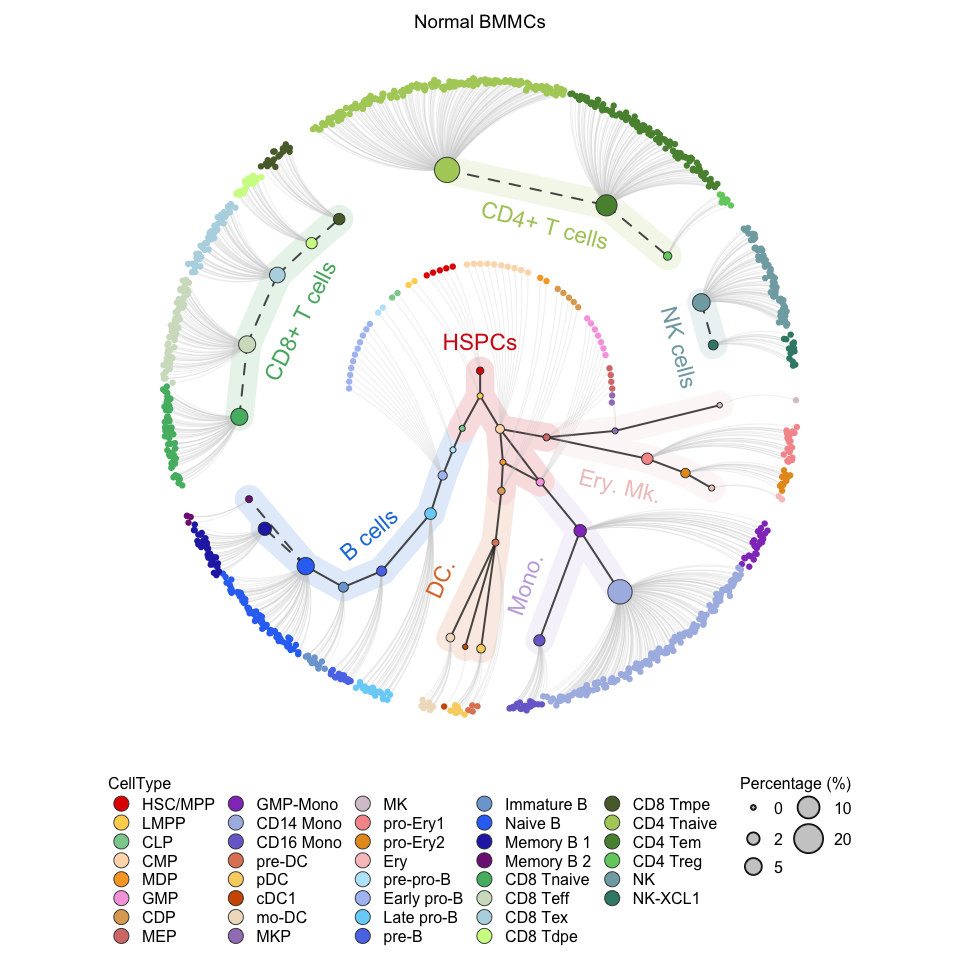
# Visualization for APL (circle tree)
plotCircleTree(hemato.subc = hmap, group.subc = "external", top.subc = 3,
color.mapping = "cell.percentage", title = "APL BMMCs")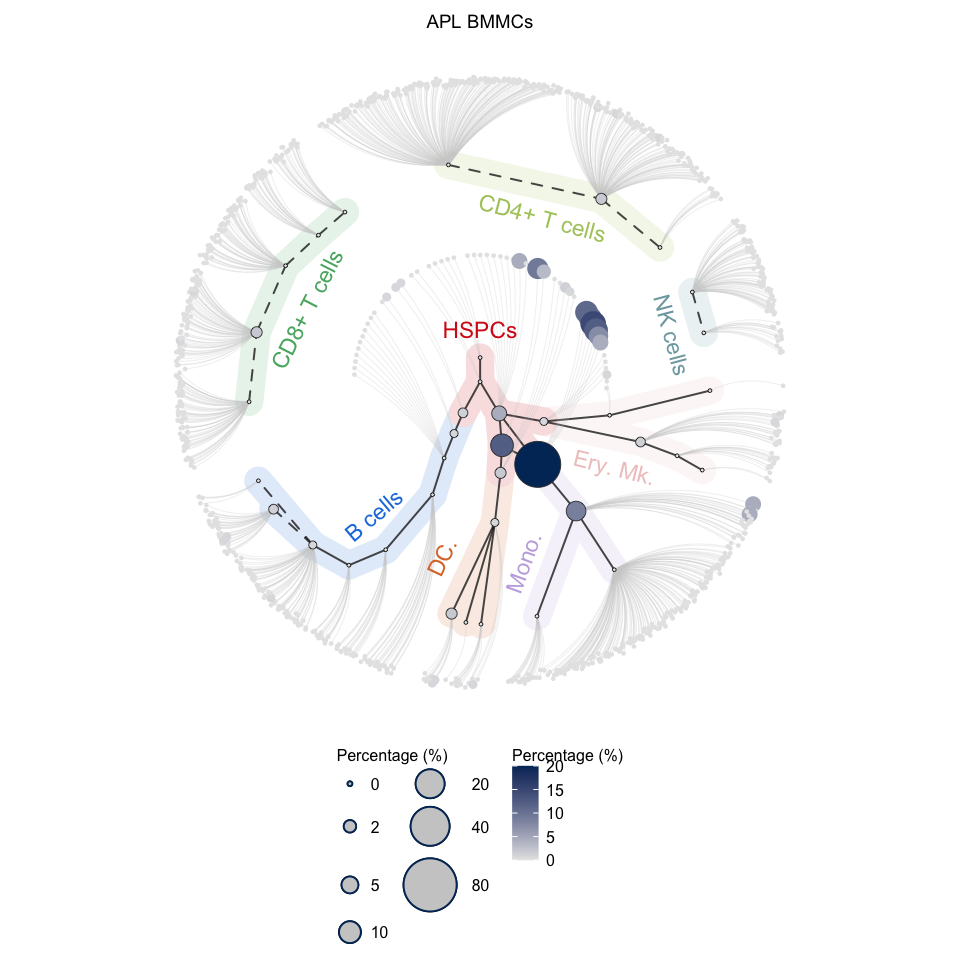
# Visualization for the normal BMMC (the reference) (circle tree)
plotCircleTree(hemato.subc = hmap, group.subc = "reference", top.subc = 3,
color.mapping = "cell.percentage", title = "Normal BMMCs")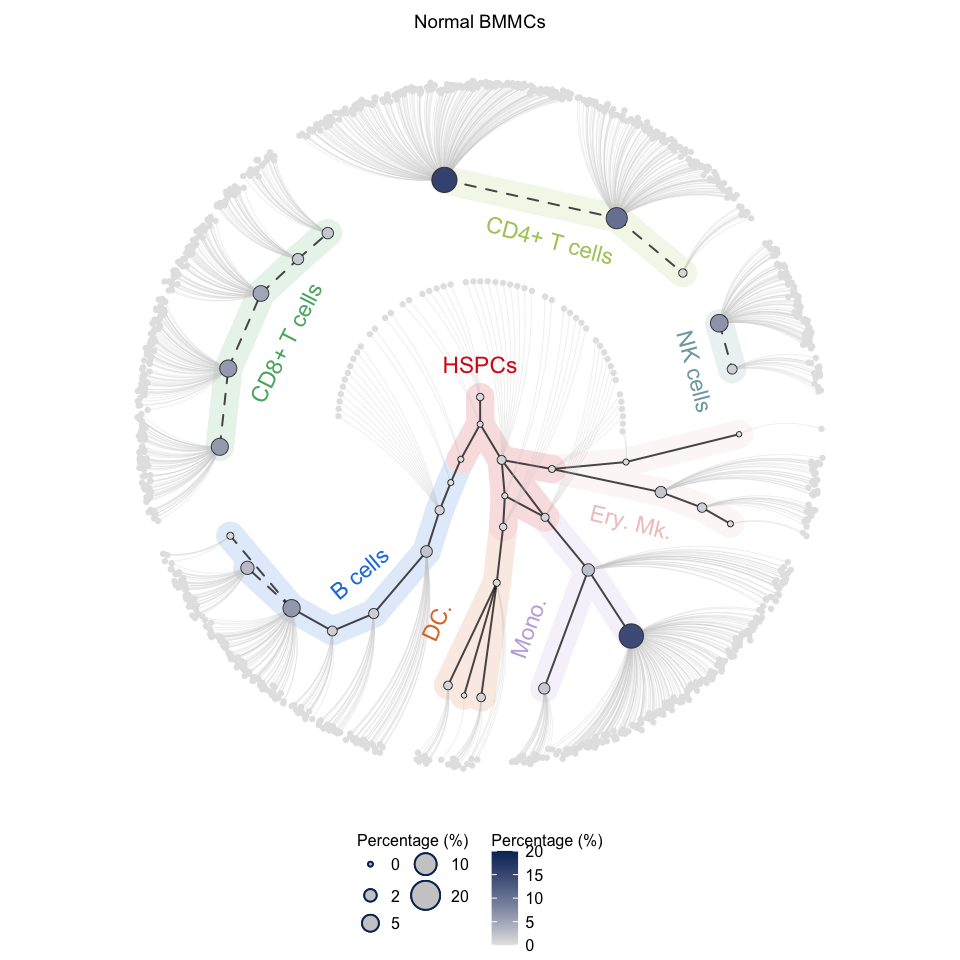
Visualization using the absolute proportional mode.
# Absolute proportional mode
# Visualization for APL (hierarchy tree)
plotClusterTree(hemato.subc = hmap, group.subc = "external",
color.mapping = "cell.type", title = "APL BMMCs",
size.mode = "absolute.pct", top.subc = 3,
point.size = 12, label.cell = TRUE)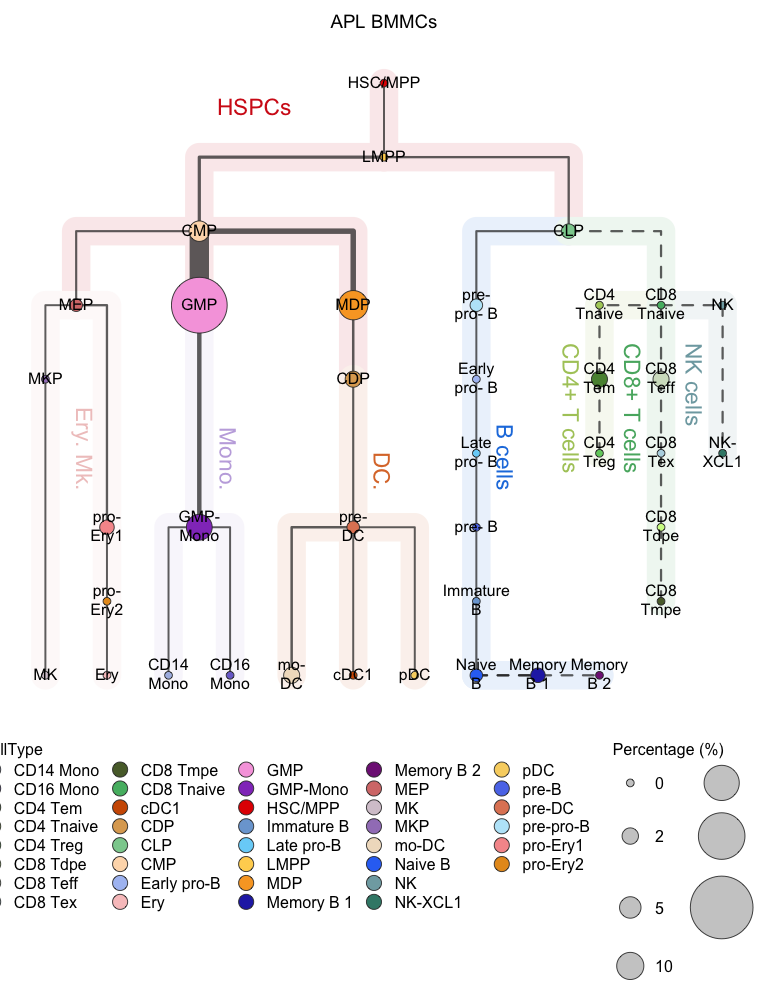
# Absolute proportional mode
# Visualization for the normal BMMCs (hierarchy tree)
plotClusterTree(hemato.subc = hmap, group.subc = "reference",
color.mapping = "cell.type", title = "Normal BMMCs",
size.mode = "absolute.pct", top.subc = 3,
point.size = 12, label.cell = TRUE)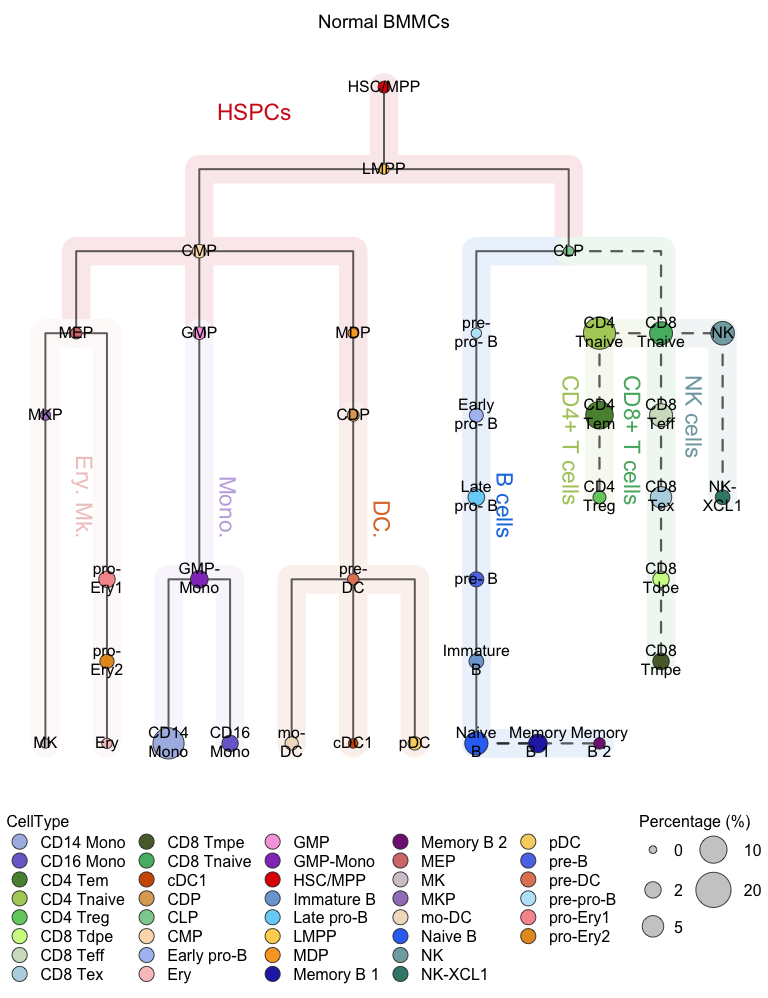
# Absolute proportional mode
# Visualization for APL (hierarchy tree)
plotClusterTree(hemato.subc = hmap, group.subc = "external",
color.mapping = "cell.percentage", title = "APL BMMCs",
size.mode = "absolute.pct", top.subc = 3,
point.size = 12, label.cell = TRUE)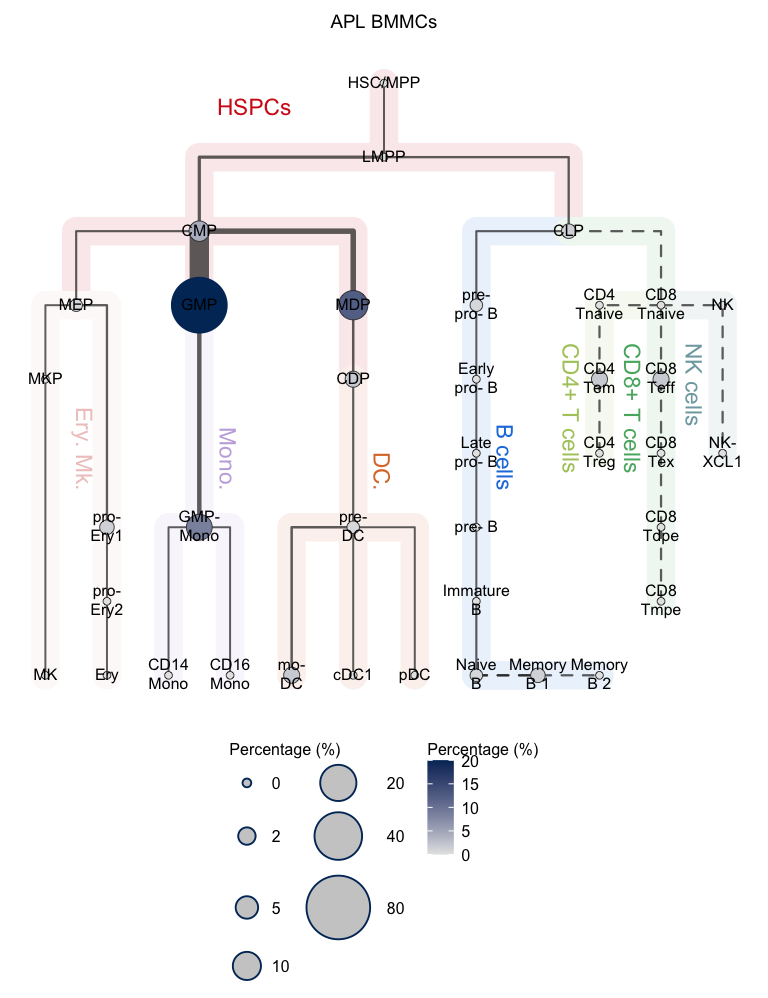
# Absolute proportional mode
# Visualization for the normal BMMCs (hierarchy tree)
plotClusterTree(hemato.subc = hmap, group.subc = "reference",
color.mapping = "cell.percentage", title = "Normal BMMCs",
size.mode = "absolute.pct", top.subc = 3,
point.size = 12, label.cell = TRUE)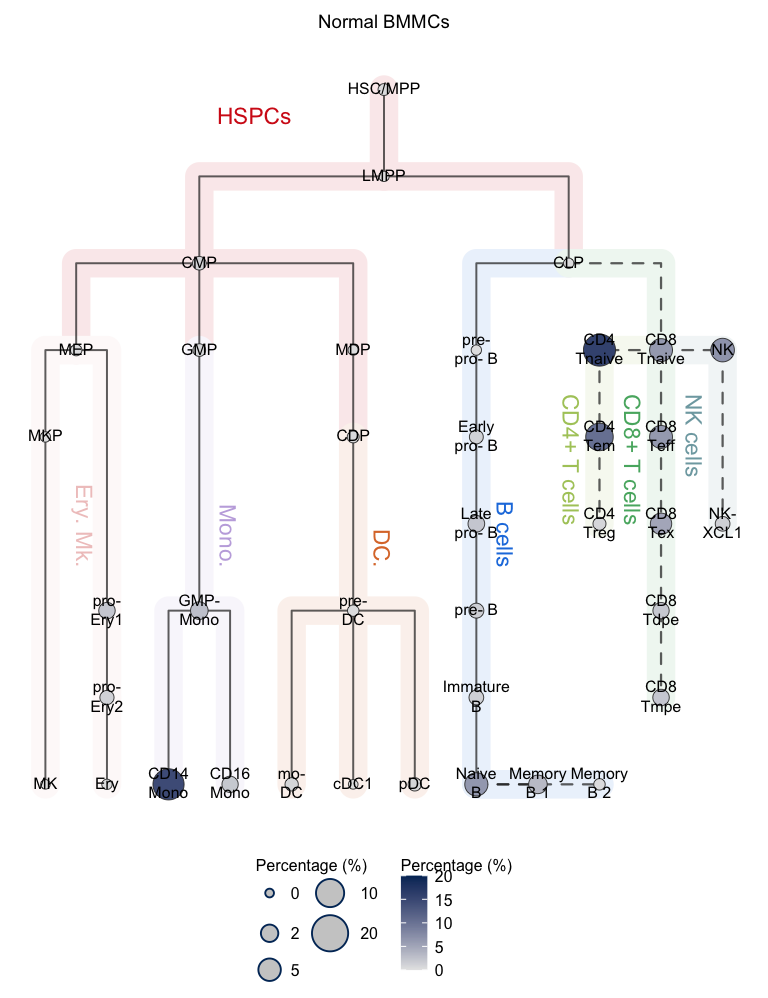
Visualization using the relative proportional mode.
# Relative proportional mode
# Visualization for APL (hierarchy tree)
plotClusterTree(hemato.subc = hmap, group.subc = "external",
color.mapping = "cell.type", title = "APL BMMCs",
size.mode = "relative.pct", top.subc = 3,
point.size = 12, label.cell = TRUE)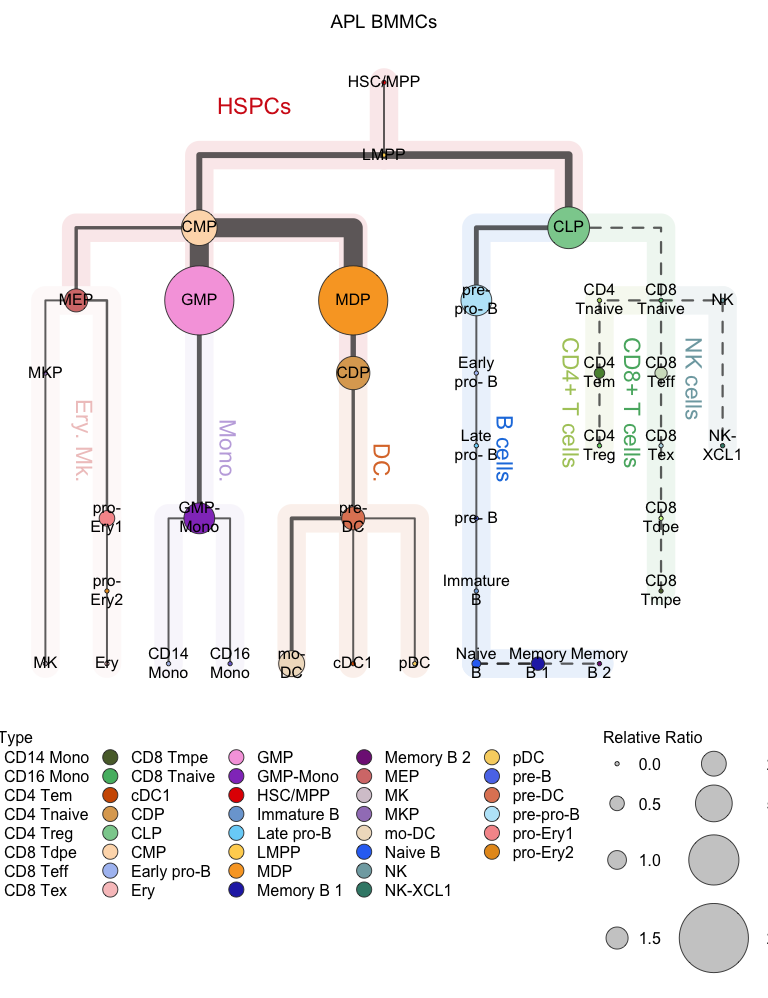
# Relative proportional mode
# Visualization for the normal BMMCs (hierarchy tree)
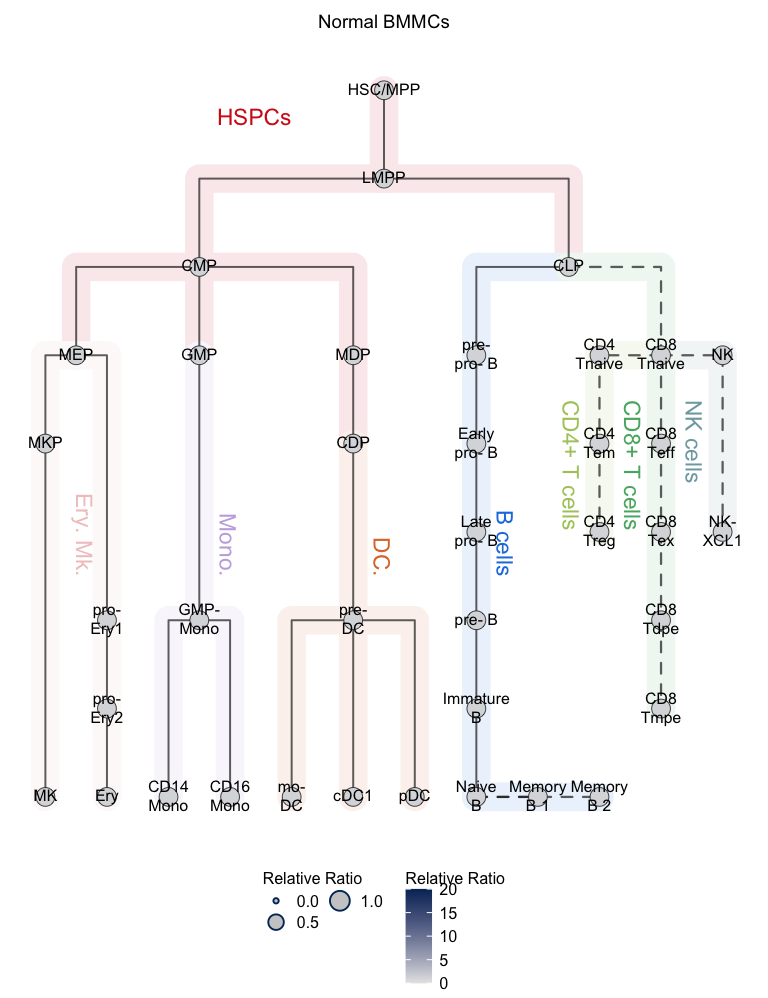
plotClusterTree(hemato.subc = hmap, group.subc = "reference",
color.mapping = "cell.type", title = "Normal BMMCs",
size.mode = "relative.pct", top.subc = 3,
point.size = 12, label.cell = TRUE)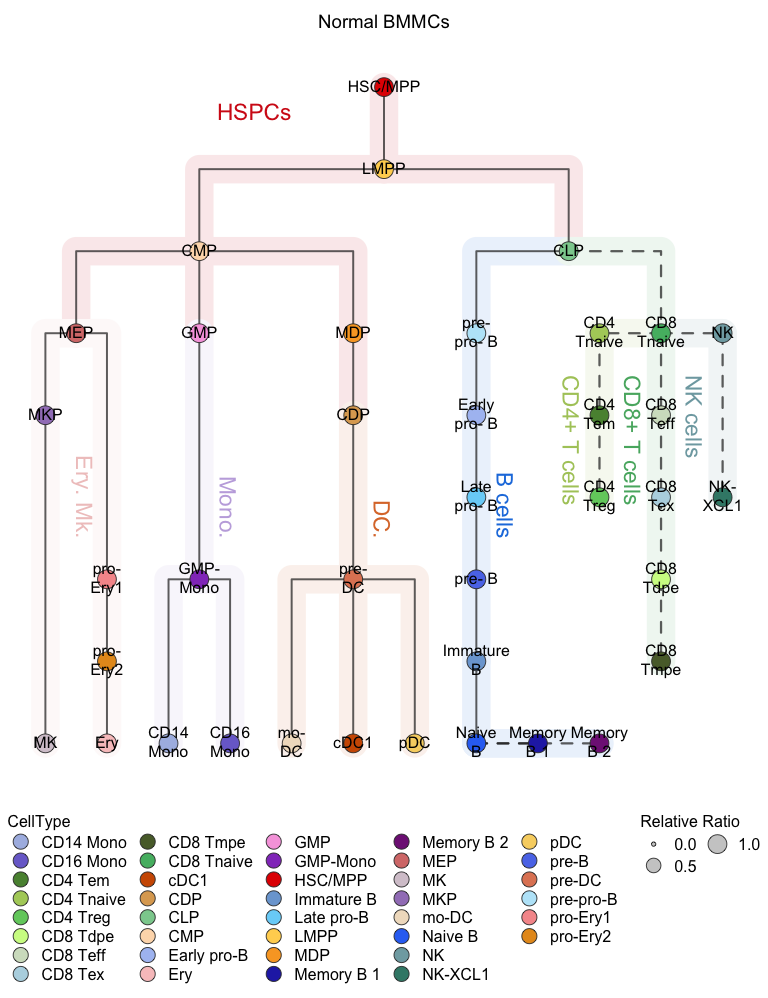
# Relative proportional mode
# Visualization for APL (hierarchy tree)
plotClusterTree(hemato.subc = hmap, group.subc = "external",
color.mapping = "cell.percentage", title = "APL BMMCs",
size.mode = "relative.pct", top.subc = 3,
point.size = 12, label.cell = TRUE)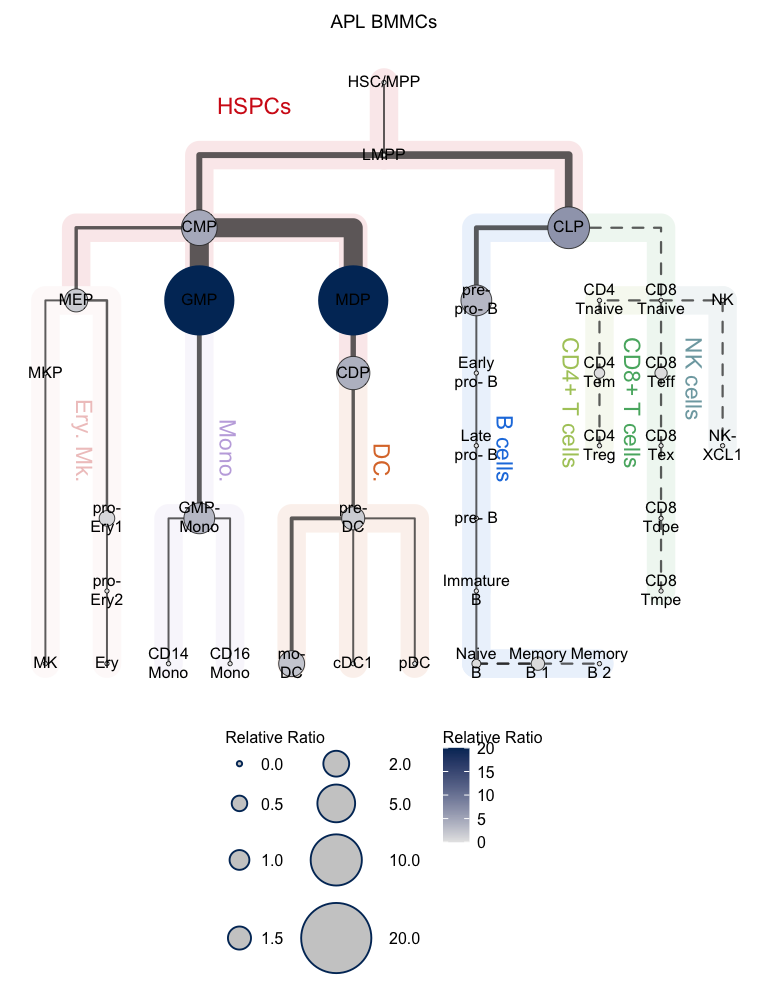
# Relative proportional mode
# Visualization for the normal BMMCs (hierarchy tree)
plotClusterTree(hemato.subc = hmap, group.subc = "reference",
color.mapping = "cell.percentage", title = "Normal BMMCs",
size.mode = "relative.pct", top.subc = 3,
point.size = 12, label.cell = TRUE)